The UCSC CSC offers two quite different imaging platforms: the Phenix and the ImageXpress.
I. PHENIX
Basic information:
-

Beverley Rabbitts and Laura Rodriguez Velandia detecting the effects of novel natural products on cells using the UCSC CSC Phenix. Make/Model: Perkin Elmer Opera Phenix Plus
- What makes it special compared to other imagers: fast, the most high end currently on the market
- Capabilities in brief: two camera spinning disk confocal microscope with 4 lasers, water objectives up to 63x, controllable imaging environment (temperature, gas, humidity) for live cell imaging, brightfield and digital phase contrast.
- For high throughput: Integrated Flex robot for automated sample loading from a plate hotel or cell incubator.
- Interface: A controller PC that uses Harmony software. It also can remote-connect to a second PC that uses PlateWorks software to control both the robot and the Phenix, calling on imaging protocols saved within Harmony.
- Confocal: Spinning discs with fixed pinhole size of 50 um (or option to image in widefield mode)
- Microplates: Please note that these objectives are optimized for a plate bottom thickness of 0.17 mm but the 20x air objective has a correction collar to accommodate 0-1.5 mm thickness. The plate bottom should be glass or imaging-quality plastic, with black walls (such as the PhenoView plates from Perkin Elmer). Or we also have a holder for classic slides.
-
Flex robot for sample loading to the Phenix
- For high throughput screening (with automated imaging)
- Samples are loaded from either a cell incubator or a room temperature plate hotel.
- The incubator has the same racks as the incubators on our Explorer workstation, and they can be interchanged so plates can be moved in batch between the workstations.
- Other special capabilities: "Preci-Scan" for zooming in on rare events after a low mag snapshot, stitching together multiple sites from one well, flatfield correction, 3D visualization of Z stacks. See the bioinformatics page to read about segmentation, quantification, and statistics built in to Harmony.
We do not currently offer:
- additional colored lasers, higher power lasers, deconvolution, heating to temperatures other than 37 degrees, optical phase contrast, different sized pinholes, simultaneous capture of two channels using the same excitation laser.
- the following that are optional packages could be acquired with your contribution:
- the onboard pipetor. If you need to add reagents before or between measurements, we suggest you do so by manually taking your microplates to our separate dispensers, robots, or use a multichannel pipet.
- up to two additional cameras are possible for optimal speed of simultaneous channels.
- 40x water objective
- Objectives currently available on the CSC Phenix:
| M | immersion medium | NA | WD (mm) | FOV width (mm) | depth of focus (um)* | r in xy (um)* | ideal z slice spacing (um)* |
|---|---|---|---|---|---|---|---|
| 10x | air | 0.3 | 5.2 | 1.29 | 16.2 | 1.3 | 7.4 |
| 20x | air | 0.4 | 8.28 | 0.65 | 8.0 | 0.79 | 3.6 |
| 20x | water | 1.0 | 1.7 | 0.65 | 1.8 | 0.66 | 0.8 |
| 63x | water | 1.15 | 0.6 | 0.21 | 1.0 | 0.28 | 0.5 |
- *theoretical values of depth of focus, xy resolution (r), and z slice spacing is based on the numerical aperture (NA) and an average wavelength.
- Working distance (WD) is the distance from the lens to the focal point (therefore it includes the width of the microplate bottom or coverslip normally excluded from WD usually published for microscope objectives).
- Please note that these objectives are optimized for a plate bottom thickness of 0.17 mm but the 20x air objective has a correction collar to accommodate 0-1.5 mm thickness.
- Lasers: four excitation laser lines (405, 488, 561, and 640 nm) and one long wavelength laser for autofocusing via two peaks detection of the plate and well bottoms.
- Detectors: Two sCMOS cameras for simultaneous imaging of two channels in confocal mode. Note that the two cameras are capturing light from spatially separated illuminated regions of the sample, so there is no crosstalk/bleedthrough (unlike for a classic point scanner's simultaneous imaging). Also note that because of this, the two cameras use separate light paths and therefore are associated with specific channels:
- one camera for 405 nm and 561 nm excitation
- 435-480, 435-550, 465-480, or 465-530 nm filters for 405 nm excitation
- 570-630, 571-596, or 605-630 nm filters for 561 nm excitation
- one camera for 488 nm and 640 nm excitation, usually used with these emission filters:
- 500-530, 500-550, or 515-550 nm filters for 488nm excitation
- 650-680, 650-760, or 690-720 nm filters for 640 nm excitation
- one camera for 405 nm and 561 nm excitation
- Therefore to take advantage of the two camera system, you must image your DAPI or TRITC at the same time as your FITC or Cy5.
- Other excitation/emisssion combinations might be possible in certain cases, and almost all combinations are possible in widefield mode. The possible combinations are listed below, with possible being defined as those that are physically capable of being lined up in the filter wheel and readily selectable in the Harmony software (Y=yes, N=no). For more details, see the filter inventory.
| imaging mode: | confocal | widefield | |||||||
| excitation laser: | 405 | 488 | 561 | 640 | 405 | 488 | 561 | 640 | |
|
nm range of emission bandpass filters
|
435-480 | Y | N | Y | N | Y | Y | Y | Y |
| 435-550 | Y | N | Y | N | Y | N | Y | Y | |
| 465-480 | Y | N | Y | N | Y | Y | Y | Y | |
| 465-530 | Y | N | Y | N | Y | N | Y | Y | |
| 500-530 | Y | Y | Y | N | Y | Y | Y | Y | |
| 500-550 | Y | Y | Y | Y | Y | Y | Y | Y | |
| 515-550 | Y | Y | Y | Y | Y | Y | Y | Y | |
| 570-630 | Y | N | Y | N | Y | Y | Y | Y | |
| 571-596 | Y | N | Y | N | Y | Y | Y | Y | |
| 605-630 | Y | N | Y | N | Y | Y | Y | Y | |
| 650-680 | N | Y | N | Y | Y | Y | Y | Y | |
| 650-760 | N | Y | N | Y | Y | Y | Y | Y | |
| 690-720 | N | Y | N | Y | Y | Y | Y | Y | |
We have Perkin Elmer Signals Image Artist bioinformatics package, Spotfire, and a second copy of Harmony software on a computer workstation in the lab space. It is currently directly networked with the Phenix imager controller PC for fast data transfer.
Computer specifications:
2 x Xeon Gold 6226R CPU w/ 16 cores each
128 GB RAM
500 GB SSD for OS
72 TB RAID5 disk array (6 x 16 TB SAS drives)
10 GbE copper NIC
Power supply:
1400W at 181VAC – 240VAC
1100W at 100VAC – 180VAC
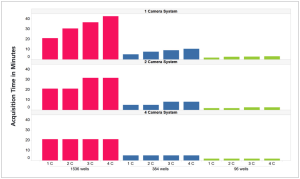
Click here to read Perkin Elmer's flyer showing how different variables influence the speed of image acquisition. One of the figures is shown below – note that we have a 2-camera system which helps make it faster for even numbers of channels.
The samples can either be slides (we have a special slide holder for that) or microplates.
Required microplate properties:
- standard size in XY
- clear bottom (they are imaged from the bottom i.e. inverted lightpath)
- sturdy hard plastic (so the robotic stage can grip the plate well)
Strongly suggested microplate properties:
- flat bottom (the autofocus has an easier job)
- black wall (reduces fluorescence noise)
- square well (so you can capture more square sites per well)
- skirt height below 0.3mm (in z direction) if you want to use 63x objective
- if the bottom is too close to the bench (no skirt rising it higher), it will scratch
- if the bottom is too high off the bench (long skirt and raised wells), the objective will crash into the skirt when it tries to get near the edge wells, and so you won't be able to image the region closest to the edge, especially a problem for short working distance objectives (to get high numerical aperture, they capture light at a wider angle, so they must be close to the plate) and water immersion objectives (because they have a collar around them maintaining the bead of water on the objective).
Optional microplate properties:
- sterile for if doing cell work in them
- tissue culture treated for cell culture in them (or polylysine or collagen coated as needed)
- lidded for convenience
- barcoded if you plan to use pick lists and/or process lists on the Echo or PlateWorks
Suggested microplates – PhenoPlate brand from Perkin Elmer.
- Here is the detailed spec sheet for the 384 well format PhenoPlate. Note that if you see Cell Carrier Ultra brand plates, these are the same product renamed.
Other resources:
- Watch a movie of me using the CSC Flex robot to load a sample into the CSC Phenix!
- Watch a promotional video by Perkin Elmer about this system.
- Phenix user manual (110 pages PDF) describes hardware, instructions for maintenance
- Phenix application guide for high content analysis of image screens (164 pages PDF)
- Harmony software manual (730 pages PDF)
II. IMAGEXPRESS
Basic information:
-

The UCSC CSC Image Xpress. Make/Model: Molecular Devices ImageXpress Micro XLS
- What makes it special compared to other imagers: has a reference database of image data collected for a decade in the CSC, is used historically in many other laboratories for similar purposes in the literature.
- Capabilities in brief: widefield fluorescence automated microscope with one camera, bright fiber optic light source, and 4 filter-based channels, and Nikon air objectives: 4x, 10x, 20x, and 40x. Operated by MetaXpress software, which also does analysis.
- For high throughput: is made to be programmed to image the whole plate with one set of settings – that’s thousands of images collected hands-free. The robotic stage that accommodates multi-well plates and slides (in a slide holder insert).
» VIDEO! of our imager in action selfie-style by Beverley Rabbitts.
» VIDEO! of MetaXpress image analysis and export selfie-style by Beverley Rabbitts.
Imaging project protocols
- EdU quantification by imaging with any cell line!
- Immune-cytological profiling in RAW and HeLa cells current (Beverley’s) summarized protocol here and longer, more detailed/complete historical (Tannia’s/Britney’s) protocols for RAW here and here or HeLa here.
- ImmuneCP measurements – historical and current
- A table to help schedule CP experiments

Other resources
- Tips on preparing figures with images, written by IBSC here.
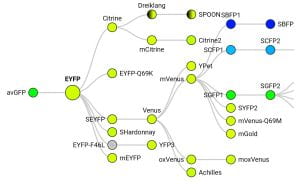
FP Base database of fluorescent proteins. It includes a diagram showing the origins of each fluorescent protein colored by emission wavelength (example below for eYFP), including relatively obscure FPs, and useful information for each variant such as the time they take to fold after translation, whether they’re a monomer, and pKa.
Some comments on image data management…
High throughput imaging lets you collect tens of thousands of high-resolution tiff images per hour! That’s up to a TB of data per day! Although we purchased significant data storage for our new imaging and image analysis computers, it could quickly get used up if we don’t manage it carefully. Here are some best practices that the CSC will be enforcing:
- When users are trained, we will communicate these issues and show you how to save your data on your own portable drive.
- We recommend that you save very valuable data in two completely separate locations (e.g. a drive at your work desk, a drive you keep at home, and in the cloud).
- For user labs with more than 1TB of data on our computer, we will charge a fee per month that it sits there.
- Hopefully this motivates users to delete anything they no longer need (false starts, outdated experiments, etc).
- Note that it is also possible to delete the raw image files but keep the quantified analysis results and metadata, which are much smaller.
- Especially if our storage space is getting full, the CSC staff will periodically delete old data. For now, we are giving you a 1 year grace period from the time that the images are collected until the data get deleted. The Phenix was installed in spring 2022 so this means nothing will get forcibly deleted until 2023 – stay tuned for announcements as this evolves.
- The university offers help with data management in several capacities.
- Click here for an online textbook called “Data Management for Researchers” at the UC Library
- Please consult Christy, the UCSC research data librarian, to learn more! including about classes she teaches on this topic.
- The UC library offers several tools and tips.
- DMPTool guides you through the process of developing a research data management plan. These are required for new NSF and NIH research grant submissions. DMPTool is offered by the California Digital Library, a division of UCOP. If you need any information during this process about the nature of the data collected by the CSC imagers, feel free to shoot us your questions. It’s in everybody’s best interests to have a good strategy in place before the data are generated. You can also request a “data consultation” by emailing research@library.ucsc.edu.
- Go to DRYAD for a place to save your already-published data where it will be publicly accessible. This service, from UC3 (University of California Curation Center), helps you meet mandates from grant agencies and publishers and also gets you a DOI for your dataset. Note that if your publications are not yet free of charge to the public, you can give them open access through UC’s open access tools and eScholarship.
- For unpublished data, you can store it through Merritt, a paid service also from UC3. And FYI all sorts of unpublished data can get a persistent unique identifier using UC’s EZID tool.
- The CSC is working on data storage solutions for larger volumes of image data. This is a major effort that involves the whole university to try and modernize UCSC’s research data storage (and networking to connect us to storage locations). For more information, you can talk with Hart Hancock and Eric Shell. We recommend UCSC researchers to notify the leadership if their needs in this area are not currently being met, so that the university will take action towards a unified structural solution. So in this case, we’re asking you to not be patient while these resources evolve!